Research Article
Ajay Nair1*, Peter K Dearden2
1 Arthritis & Clinical Immunology Research Program, Oklahoma Medical Research Foundation, USA
2 Department of Biochemistry, University of Otago, New Zealand
Corresponding author
Ajay Nair, Arthritis & Clinical Immunology Research Program, Oklahoma Medical Research Foundation, Oklahoma City 73104, Oklahoma, USA, Tel: 1 (405)985-6960; E-mail: ajay-nair@omrf.org; Received Date: 15th November 2016; Accepted Date: 21st December 2016; Published Date: 26th December 2016
Citation
Nair A, Dearden PK (2016) Waddington?s Assimilation, A Fact or A Mere Philosophy Shaped in The Lamarckian Mold: A Genomic Inquiry. Enliven: Bioinform 3(1): 001.
Copyright
@ 2016 Dr. Ajay Nair. This is an Open Access article published and distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.
Abstract
Wadington’s genetic assimilation experiment, in 1953, was one of the first experiments that claimed the inheritance of an acquired character. In this experiment, he applied a heat shock to flies triggering a low frequency of a “crossveinless” (cve) phenocopy (posterior crossvein defects in the adult wing). Through repeated selection of this trait with heat-shock, he was able to fix cve in the population. Waddington also believed that if the classic experiment were to be repeated with a different foundation stock, the same phenotypic effect might be produced. Microarray analysis comparing RNA expression in the wing imaginal discs of heat-shocked pupae between two laboratory fly stocks (wild-type canton S flies and w1118 strain) identified novel genes that were differentially expressing in the w1118 strain. Thus showing that both strains differ considerably in their capacities for cve production Resequencing the whole genome suggests that cve alleles are common, naturally occurring polygenes spread on all three major chromosomes of Drosophila, and that they act additively to produce wings with disturbed posterior crossveins. Widespread position effects in the genome alter key epigenetic and developmental processes to produce the phenocopy.
Introduction
The fate of a developing trait is plastic, which provides a channel for rapid evolution [1]. This argument is based on the fact that selection can act on ancestral developmental pathways to produce alternate forms of a phenotype; thereby lead to evolutionary divergence (?genetic accommodation?; [2]). However there are cases when plasticity may become reduced or lost from repeated selection, and an induced trait might get fixed or ?assimilated? without the original environmental cue [3]. Waddington established the concept of canalization to explain how the wild form of an organism remains consistent and less variable towards external disturbances [4]. To draw parallels between canalization and the manifestation of cryptic traits, Waddington showed that certain phenotypes (for e.g. cve; [5]) could be produced at a low rate in a population by heat stress. He went on to select cve flies from heat-shocked populations, and showed an increase in the rate of cve. After x generations of selection, however, not only had the novel trait reached high levels, but heat-shock seemed no longer necessary to induce it. Waddington explained this apparent assimilation of an acquired character as the effects of selection on the activation threshold for the phenocopy in question[5-11]. In other words, as long as genetic variation exists, mechanisms that dampen the effects of variation on that trait are likely to get favored by stabilizing-selection [12]. Despite the experimental and theoretical underpinning laid by Waddington, the mechanisms and evolutionary causes of genetic assimilation remain obscure. Recently canalization has gained much more attention; particularly with respect to Hsp90 as a capacitor that stores and releases genetic variation under external stress [13,14]. The presence of phenotypic deviants whose expression depends on genetic buffers (such as Hsp90) has far-reaching implications for the ability of selection to affect phenotypic plasticity and evolvability.
We report here the results from Microarrays and Whole Genome Resequencing following a successful repetition of Waddington?s genetic assimilation experiment ([15], Figure 1). Here we show that the rarity of crossveinless in the wild could be seen as evidence of low frequency alleles, spread across the genome, sensitive to environmental effects on penetrance and viability. Group of differentially expressing novel genes that might be misfiring in producing the stress induced trait.
The process of artificial selection rather than random, de novo mutations allow cve alleles to be selected, accumulated and passed on to future generations. The results, therefore, not only indicate a polygenic basis for crossveinless but also imply that this stress induced trait affects the fly on a broader scale than it was understood before.
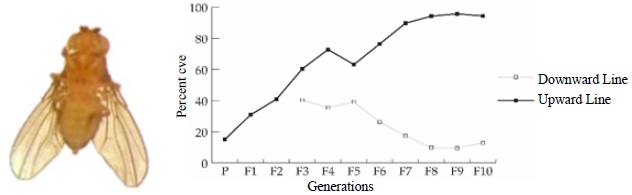
Figure 1. Response to selection, in ten generations, for crossveinless wings (?upward? selection) and normal wings (?downward? selection) in the Waddington experiment. (left) w1118 strain with disturbed poseterior crossveins in one of its wings. (right) Line graph presenting an overall increase and decrease in the percentage of crossveinless over ten generations in the Upward and the Downward Selection Line (Nair A., Dearden P.K., 2016).
Results and Discussion
Hsp90 Does not Explain the Strain Specific Nature of Crossveinless
One likely explanation for the strain specific nature of our crossveinless trait is that it reflectscryptic genetic variation that is normally repressed by the actions of capacitors, like Hsp90 [6,14]. To test if disruption of this capacitance induces the expression of crossveinless, we used Geldanamycin (GA) [16] to specifically block the activity of Hsp90. By raising flies from both the stocks (the Canton-S flies and the white-eyed w1118stock) in geldanamycin treated media and examining for crossveinless, we hoped to determine if the crossveinless trait is buffered by Hsp90, and reflects cryptic genetic variation present in thew1118, but not the Canton-S flies. Despite observing a number of other defects, neither fly line showed any crossvein defects (Supplementary - S1 - Table 25, Table 26). This implies that Hsp90 is not the key factor buffering heat-induced cve. There were a couple of flies with the c-like phenotype in the DMSO control groups (Supplementary - S1 - Table 26) giving an impression that the effect might be produced due to the solvent alone, and not as a result of geldanamycin mediated hsp90 inhibition. But it has been shown that use of DMSO might complicate mutagenicity screens by giving false positive results.
Novel Retrogenes differentially Expressed between Heat-Shocked and Non-Heat-Shocked Wing Discs
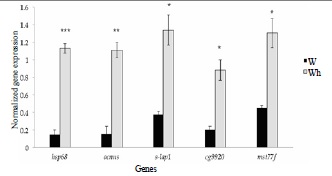
To understand possible causes for the cve phenotype, we examined RNA expression in the wing imaginal discs of non heat-shocked and heat-shocked pupae of both w1118 and Canton-S. By comparing gene expression in wing discs of our susceptible strain with those from non-susceptible strains we aimed to identify genes whose expression may differ in reponse to heatshock between the strains. A more comprehensive study, using whole genome gene expression arrays (Affymetrix Inc., CA, USA), reported a detailed analysis of the full heat stress response in Drosophila melanogaster females. The study looked at both up- and downregulated genes just before and after an application of heatshock (36°C for an hour). Three heathsock response patterns were observed in three independent clusters and these were named as: Early-up, Early-downregulated and Late-up. Early-up included genes known to be heat responsive and to belong to the group of genes having chaperone activity. The genes found to be early-downregulated and late-up by heatshock mainly related to metabolism. In order to draw comparisons, heat-shocked samples were grouped with non heat-shocked samples. Out of 2900 Gene Ontology (GO) terms enriched from the GO analyses on the group Wild type (Wt) vs. Wild type-heat-shocked (Wth), statistically significant genes (with FDR-corrected p-value < 0.05 and fold change >±2) included those concerned with processes like: response to stress, heat-shock mediated polytene chromosome puffing, response to heat (Supplementary - S2). Most of the genes in this group were those expressing Hsp70 family of proteins. The White (W) vs White-heat-shocked (Wh) group however, amongst the expected responses to heat, as seen in Canton-S flies, also expressed a set of genes that were not found in the canton-s dataset. Many of these genes we identified as being unusual because they are believed to be testis enriched (Supplementary - S2). The expression of a subset of these novel genes was validated using Q RT-PCR (FFigure 2,Supplementary - S3). Although these genes have been known to be testis specific/enriched, many are either novel retrogenes (like cg9920andocnus) or are functional elements of the Drosophila heterochromatin (like mst77f and s-lap1, [17]). Most of these retrogenes known to be testis related are known to be involved in chromatin remodeling and position effects in the Drosophilagenome [18].
Figure 2. Column graphs showing the differentially expressing genes from the W vs. Wh group that were validated using Q RT-PCR. The primary y-axis (green and red columns) consists of average expression mean values obtained by normalizing real-time gene expression values from individual triplicates in the W vs. Wh group. Statistical significance was estimated by first measuring the standard deviation and then the standard error mean from the individual means. Validated genes (x-axis) were statistically significant (p-values * <0.05; ** <0.005; *** <0.0005) with fold changes >2.
Whole Genome Re-sequencing: Clues to the cveComplex
The microarray data implied that differences in response to heatshock may underly the development of the crossveinless phenotype. That a collection of functionally related genes is expressed in the susceptible stain, but not Canton-S, perhaps indicates that variations in the genome of these two strains may explain their different responses. To determine if this is the case, and if these variations are enriched in the assimilated lines, we resequenced whole genomes from three strains of Drosophila melanogaster: the wild-type strain (Canton-S, Wt), the white-eyed (w1118) strain and the assimilated line (GA) generated during our repeat of the Waddington experiment.
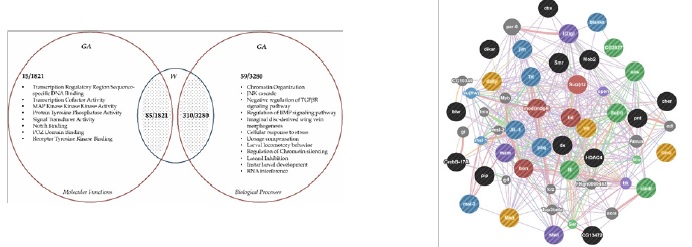
Upon analyzing the genome data, it was interesting to see that significant variants (with coverage> 10; forward/reverse balance> 0.22; p<0.05) in the assimilated sample were spread across three major chromosome arms (2,3 and X). From the Gene Ontology (GO) Enrichment analysis, three biological processes were seen consistently, with high statistical significance (p<0.001), across the major chromosomes: regulation of chromatin silencing, chromatinorganization and lateral inhibition (Supplementary - S1, Table 29).The three processes mentioned above are basal processes that could regulate large numbers of genes, developmental processes, as seen in the gene expression data. Out of 3280 biological processes and 1821 molecular functions compared between the samples W and GA, 59 biological processes and 15 molecular functions were significantly over or under represented in GA flies (Figure 3a,Supplementary - S1, Table 29, Table 30).
Some of the key variant genes identified were: chromatin modifier gene products (such as Mod(Mdg4), JIL-1, Trletc; Figure 3b) known to be part of an active mechanism that provides means to compartmentalize the genome and prevent heterochromatic spreading into active euchromatic regions; loss of function alleles have been shown to act as enhancers of variegation (Sass G.L., 1998). starvin is the Drosophila member of the BAG (mammalian BcII-associated athanogene) family of genes that is key to larval development in many ways, such as: stress response, locomotion, feeding and survival. starvin mutant larvae fail to properly grow and crawl after hatching. In several instances larval growth remained arrested in the first instar. Also, their ability to uptake food is severely impaired [19]. Feeding and Larval Locomotory behavior were one among the significantly enriched processes in the GO analysis on sample GA (Supplementary - S1, Table 29). Also, compromised pupal viability and pupation closer to the food source displayed by cve individuals during the repetition of the Waddington experiment [15] could be due to their inherent inability to uptake food and crawl well.
Figure 3. (a) Venn diagram illustrating significant biological processes and significant molecular functions from the assimilated sample, following GO enrichment analysis. (b) Gene network prediction on annotated genes using GeneMania. Six main types of networks are depicted: Co-expression ( ), Physical interactions ( ), Co-localization ( ), Shared Protein Domains ( ), Predicted ( ) and Genetic interaction ( ). Grey circles indicate novel genes and interactions observed in the network that were not submitted in the initial default query. Blue circles indicate genes and interactions involved in the regulation of chromatin silencing. Red circles indicate genes and interactions involved in chromatin organization. Green and Purple circles indicate genes and interactions involved in lateral inhibition or determination of cell fate. Yellow circles indicate genes and interactions involved in imaginal disc derived wing-vein morphogenesis. Black circles indicate genes and interactions observed in the network that were involved in other developmental processes. Grey circles indicate novel genes and interactions observed in the network that were not submitted in the initial default query.
However it was interesting to see that the changes enriched in the GA lines were present in the w1118 read dataset at low frequencies. This means that the genetic assimilation process is selecting for these alleles, rather than this being a result of random de novo mutations. Also, upon comparing w1118 variants with the Canton-S read data, it became apparent that the distribution of the read positions variant in the w1118 variants carrying reads is different from that of all the reads covering the variant position (read-position test probability (p<0.05); Supplementary - S1, Table 31).This could explain why the White-eyed strain was susceptible to cve initially.
Conclusions
Repeating Waddington?s genetic assimilation experiment [15] lead to an interesting discovery. Among the two laboratory Drosophila strains that were used in the experiment, the wild-type (the wild Canton-S) failed to produce disturbed posterior crossveins and the white-eyed strain (the white-eyed mutant w1118 strain) succeeded in not only producing but also allowing the trait to be selected for in the following generations. How is it that despite having common predisposing factors, and being reared under identical conditions, the two strains responded differently to the stress response? Clearly, as stated earlier, the presence of minor modifier genes may well make stains differ from each other in terms of their physiological response to stress.
By comparing transcriptomic data from the two laboratory strains used in the present replication of the Waddington experiment, we conclude that susceptibility towards cve is affected by unusual activation of novel genes that cause changes in gene regulation. Studying the pattern of expression of some of these novel retrogenes, a forward idea called the- ?out of the testes? theory [19] has been suggested. According to which, functional retrogenes or heterochromatin elements despite being expressed in the testis may acquire higher and broader tissue expression, ultimately leading to newer functions. From the microarray data we hypothesize that perhaps the packaging of DNA in the wing discs is different between the Canton-S and w1118 in relation to cve. This implies that an underlying difference in the w1118 genome leads to changes in chromatin organization which ultimately leads to the expression of the phenotype.
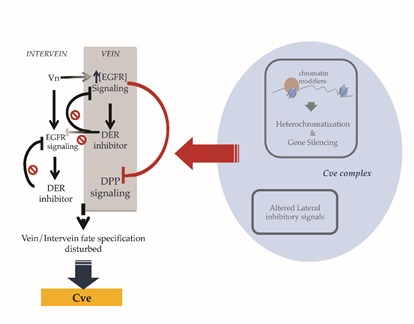
Our studies on the heat-shock induced crossveinless phenotype at the genome level allowed us to propose a model for the production of crossveinless in Waddington?s experiment. The heat-shock conditions used in the Waddington experiment seemed to produce widespread effects in the Drosophila genome. One of these changes involved chromosomal rearrangements in chromatin modifier genes, especially those that are known to cause position effects via long-range heterochromatization (for e.g. E(var) genes) in the euchromatic portions of the genome. Chromatin silencing such as these, might alter lateral inhibitory signals which are key to specifying vein/intervein cell fates during wing development (for e.g. key targets of the Su(H):NICD complex act as repressors of wing vein formation in the interveins; [20,21]). Chromatin modifiers along with lateral inhibitors perhaps constitute the ?cve complex?. GO processes like Imaginal disc-derived wing vein morphogenesis, Bone Morphogenetic Protein (BMP) signaling and Mitogen activated Protein Kinase KinaseKinase (MAPKKK) activity, enriched in the assimilated whole genome dataset, are key to the development of wing veins in Drosophila. Members of the Epidermal Growth Factor Receptor (EGFR) signaling relay wing-vein expression via MAPK activity [22,23]. Usually, in pupae 24-30 hours a.p., the expression of Drosophila EGFR (DER) inhibitors (such as argos (aos)) not only suppresses the expression of EGFR in veins but also diffuses to the interveins and suppresses DER expression activated by vein (vn). This suppression helps in properly determining vein/intervein cell fates. So it could be assumed that altered lateral inhibitory signals perhaps affect the expression of DER inhibitors. As a result, DER inhibitors are unable to reduce the overall level of EGFR signaling in the vein stripes and in the intervein region 24-30 hours a.p. Persistent EGFR signal in vein territories suppresses BMP/Decapentaplegic (DPP) signaling required for crossvein specification [23]. This inhibition could correlate with the loss of crossveins. Posterior crossvein defects seen in the current study and in the original work [5], could be either due to an inability to express crossveins or due to overexpression of interveins in regions destined to specify crossveins (Figure 4).
Figure 4: Model for the production of crossveinless during wing development in Drosophila. Long-range heterochromatization in the Drosophila genome alter notch regulated lateral inhibitory signals key to establishing vein/intervein cell fates during the development of wing veins in early pupae. A disturbed vein/intervein balance might produce crossveinless in the developing wing.
In our previous work, we have explicitly tested that (a) Flies can be selected. (b) Although cve appears without heatshock, individual fitness is compromised [15]. This work further digs into the crossveinless genome and says that there are many variants in pathways associated with crossveins. However these variants are not de novo, they exist as low frequency alleles in the population. Selection additively increases their representation in the population to produce crossveinless. Collectively we hypothesize that assimilation of cve might not be a magical incorporation of the trait in the genetic makeup but a mere outcome of strong selection.
Experimental Procedures
Origin of Experimental Flies
Induction of Heatshock and the Waddington experiment:
Drosophila prepupae were collected 120 hours after egg laying (AEL) in vials. Prepupae were collected over 12-15 time points in a day. And in order to make it possible to gather enough pupae the specific collection lasted for almost a week. Vials containing pupae were incubated for 24 hours (at 25°C) and then heat-shocked at 40.5°C for 45min in a programmable water bath (Contherm Scientific Ltd., 350-380 Series of high Temperature Digital Water Bath). Following heat-shock, the adult flies eclosed in 4-5 days, at which point their cross veins were scored.
Maintenance Regimen
Two strains of Drosophila melanogaster wereused in the current study: Wild-type Canton-S flies from Bloomington Drosophila stock center at Indiana University (BDSC) and a white-eyed w1118 stock from the same source. Fly lines were maintained on a standard cornmeal/yeast/agar medium.
Preparation of Geldanamycin treated Drosophila Media
Flies from both stocks (Wild-type Canton-S flies and the w1118) were treated with geldanamycin as described [13]. Following treatment, the progeny were scored after 10 days. In addition, to check if varying concentrations of geldanamycin resulted in any degree of severity, flies were exposed to three different concentrations of geldanamycin (1x, 0.5x, 3x).
Imaginal Wing Disc Dissection
Wing discs from Drosophila pupae were dissected roughly 24 hours after pupation. Prior to dissection, half of the pupae were heat-shocked under conditions typically used in the current repetition of the Waddington experiment. The other half was left untreated. Heat-shocked pupae were dissected immediately after the heat treatment.
Isolation of Total RNA from the Wing Discs of Drosophila melanogaster
Total RNA was isolated using TRIzol® reagent (Invitrogen Life Technologies TRIzol® manual), and later purified using the RNeasy Mini Kit (Qiagen; catalogue no. 74104) as per manufcturere?s instructions. RNA integrity was checked using using Qubit® 2.0 Fluorometer (www.lifetechnologies.com).
Microarray Hybridization and Scanning
Double stranded complementary-DNAs were prepared from the isolated RNA samples using the Invitrogen VILO kit (www.lifetechnologies.com). Approximately 250ng of total RNA was then used as a substrate to generate biotinylated cDNA according to the standard Affymetrix protocol (www.affymetrix.com/support/manuals). A total of 12-affymetrix gene chips were analysed which included three biological replicates for each experimental condition and control (heatshock vs. non-heatshock in both white-eyed and wild type strains). GeneChips were prehybridised with the hybridization buffer as per manufacturer?s instructions (UnihybTM; www.arrayit.com). Data obtained from GeneChip® (GeneChip® Fluidics Station 450) scanning was further scrutinized to identify genes that are differentially expressed across all the four samples- white (W), white heat-shocked (Wh), wild-type (Wt), wild-type heat-shocked (Wth).Data was analysed using CLC Genomics Workbench (CLC Genomics Workbench version 7).
Statistical Analysis
Raw data was normalized using RMA (Robust Multichip Average) express, [24]. CLC Genomics Workbench (CLC Genomics Workbench version 7) was used to analyze normalized expression data. An inbuilt T-test analysis tool, based on a Gaussian distribution was used to compare the mean expression level between experimental groups and then evaluate the importance of the differences relative to the spread of the data within the groups. Using inbuilt Gene Ontology (GO) tools statistically significant genes from individual sample groups were annotated further to establish a functional link between genes and heat exposure.
Quantitative Reverse Transcriptase - Polymerase Chain Reaction (Q RT-PCR)
Q RT-PCR was used to validate the results obtained from the microarray experiment. The microarrays were validated on duplicate biological replicates using the CFX96 (BioRad) systems, according to manufacturer?s instructions (Supplementary - S3).
Illumina MiSeq 2X250 Base Pair End (PE) Whole Genome Re-Sequencing
Genomic DNA (gDNA) was extracted from three strains of Drosophilamelanogaster: wild-type strain (the wild-type Canton-S flies from Bloomington Drosophila stock center; Wt), white-eyed strain (the local laboratory white-eyed w1118 stock; W), and the assimilated line (GA) developed from the white-eyed strain. High quality gDNA was extracted using the DNeasy® Blood & Tissue kit (http://www.qiagen.com/DNeasy Blood & Tissue kit//) according to the manufacturer?s instruction. DNA quality and integrity was checked using the Qubit® 2.0 Fluorometer (Life Technologies; Table 1, Supplementary - S4). Raw sequence data (Fastq sequences) were run through a standard pipeline process to check for quality. Each of the Fastq sequences were mapped against the PhiX genome. Sequences mapping to PhiX were removed from the resultant SAM file, and the fastq file was reconstructed using the Sam-Fastq.jar program from the picard suite (http://picard.sourceforge.net/; Supplementary - S4).
Re-Sequencing Analysis
Re-sequenced data was analyzed using the CLC Genomic Workbench (CLC Genomic Workbench version 7; www.clcbio.com). The typical workflow involved mapping the reads to the reference genome, variant detection, and interpretation of the variants found. Following, a Fixed Ploidy Variant detection task was launched for all the samples (Wt, W and GA). The tool reported a number of known or unknown, homozygous or heterozygous variants in the form of Single Nucleotide Variants (SNVs), Multiple Nucleotide Variants (MNVs), insertions, deletions and replacements. Later two screening tasks were launched: in the first screening, W-variants were compared to Wt- variants to filter variations specific to W. The second set of screening filtered out statistically significant GA variants (with coverage> 10; forward/reverse balance> 0.22; p<0.05) from the W variants. The inbuilt GO enrichment tool was used to examine candidate variants or their corresponding altered genes for a common functional role.
Acknowledgements
I would like to thank GRAVIDA, New Zealand and the University of Otago for providing funding and research facilities that was key to the successful completion of this work.
References
- Scoville AG, Prefender ME (2010) Phenotypic plasticity facilitates recurrent rapid adaptation to introduced predators. Proc Natl Acad Sci U S A 107: 4260-4263.
- Crispo E (2007) The Baldwin effect and genetic assimilation: revisiting two mechanisms of evolutionary change mediated by phenotypic plasticity. Evolution 61: 2469-2479.
- Waddington CH (1956) Genetic assimilation of the bithorax phenotype. Evolution 10: 1-13.
- Waddington CH (1942) Canalization of development and the inheritance of acquired characters. Nature 150: 563-565.
- Waddington CH (1953) Genetic assimilation of an acquired character. Evolution 7: 118-126.
- Buskirk VJ, Steiner UK (2009) The ?tness costs of developmental canalization and plasticity. J Evol Biol22: 852-860.
- Amzallag GN (2000) Canalization as a non-genetic source of adaptiveness during morphogenesis: experimental evidence from analysis of reproductive development in Sorghum bicolor. Biosystems 57: 95-107.
- Queitsch C, Sangster TA, Lindquist S (2002) Hsp90 as a capacitor of phenotypic variation. Nature 417: 618-624.
- Sangster TA, Bahrami A, Wilczek A, Watanabe E, Schellenberg K, et al. (2007) Phenotypic diversity and altered environmental plasticity in Arabidopsis thaliana with reduced Hsp90 levels. PloS One 2: e648.
- Salathia N, Queitsch C (2007) Molecular mechanisms of canalization: Hsp90 and beyond. J Biosci 32: 457-463.
- Milton CC, Ulane CM, Rutherford S (2006) Control of canalization and evolvability by Hsp90. PLoS One 1: e75.
- Wagner A (1996) Evolution (Lawrence, Kans) 50: 1008-1023.
- Rutherford SL, Lindquist S (1998) Hsp90 as a capacitor for morphological evolution. Nature 396: 336-342.
- Rutherford S, Hirate Y, Swalla BJ (2007) The Hsp90 Capacitor, Developmental Remodeling, and Evolution: The Robustness of Gene Networks and the Curious Evolvability of Metamorphosis. Crit Rev Biochem Mol Biol 42: 355-372.
- Nair A, Dearden PK (2016) A Systems View of Waddington?s Genetic Assimilation. Int J Bioinfo Biol Systems 1: 10-17.
- Bedin M, Gaben AM, Saucier C, Mester J (2004) Geldanamycin, an inhibitor of the chaperone activity of HSP90, induces MAPK-independent cell cycle arrest. Int J Cancer 109: 643-652.
- Brodberg RK, Mitchell MJ, Smith SL, Woodruff RC (1987) Specific reduction of N,N-dimethylnitrosamine mutagenicity in Drosophila melanogaster by dimethyl sulfoxide. Environ Mol Mutagen 10: 425-432.
- Karpen GH, Spradling AC (1992) Analysis of subtelomeric heterochromatin in the Drosophila minichromosome Dp1187 by single P element insertional mutagenesis. Genetics 132: 737-753.
- Marygold SJ, dos Santos G, Urbano J-M, Antonazzo G, Matthews BB, et al. (2014) The FlyBase Consortium (2014)
- Coulson M, Robert S, Saint R (2005) Drosophila starving encodes a tissue specific BAG-domain protein required for food uptake. Genetics 171: 1799-1812.
- Vinckenbosch N, Dupanloup I, Kaessmann H (2006) Evolutionary fate of retroposed gene copies in the human genes. Proc Natl Acad Sci U S A103: 3220-3225.
- Hasson P, Egoz N, Winkler C, Volohonsky G, Jia S, et al. (2005). EGFR signaling attenuates Groucho-dependent repression to antagonize Notch transcriptional output. Nat Genet 37: 101-105.
- Hasson P, Paroush Z (2006) Crosstalk between the EGFR and other signalling pathways at the level of the global transcriptional corepressor Groucho/TLE. Br J Cancer 94: 771-775.
- Sturtevant MA, Roark M, Bier E (1993) The Drosophila rhomboid gene mediates the localized formation of wing veins and interacts genetically with components of the EGFR signaling pathway. Genes Dev 7: 961-973.
- Martn-Blanco E, Roch F, Noll E, Baonza A, Duffy JB, et al. (1999) A temporal switch in DER signaling controls the specification and differentiation of veins and interveins in the Drosophila wing. Development 126: 5739-5747.
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, et al. (2003) Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 31: e15.
- Milkman RD (1960) The genetic basis of natural variation. II. Crossveins in Drosophila melanogaster. Genetics 45: 377-391.
- Milkman RD (1962) Temperature effects on day old Drosophila pupa. J Gen Physiol45: 777-799.
- Milkman RD (1964) The genetic basis of natural variation. I. Selection for Crossveinless polygenes in new wild strains of Drosophila melanogaster. Genetics 50: 625-632.