Original Article
Quantitation of Amino Acids in Human Hair by Trimethylsilyl Derivatization Gas Chromatography/Mass Spectrometry
Ayat H. Bani Rashaid, Glen P. Jackson, and Peter de B. Harrington*
Department of Chemistry and Biochemistry, Ohio University, Athens, Ohio 45701, USA
Corresponding author
Prof. Peter de B. Harrington, Center for Intelligent Chemical Instrumentation, Clippinger Laboratories, Department of Chemistry and Biochemistry, Ohio University, Athens, Ohio 45701, USA, Tel: +01 740 994 0265; Fax: +01 740 593 0148; E-mail : peter.harrington@ohio.edu
Received Date: 17 July 2014
Accepted Date: 08 Aug 2014
Published Date: 12 Aug 2014
Citation
Bani Rashaid AH, Jackson GP, Harrington PDB (2014) Quantitation of Amino Acids in Human Hair by Trimethylsilyl Derivatization Gas Chromatography/Mass Spectrometry. Enliven: Bio Anal Techniques 1(1): 002.
Copyright
@ 2014 Dr. Peter de B. Harrington. This is an Open Access article published and distributed under the terms of the Creative Commons Attribution License, that permits unrestricted use, distribution and reproduction in any medium, provided the original author and source are credited.
Abstract
The distribution of amino acids in hair can divulge information regarding the health (e.g., diabetes) and provide a means for detecting the history of the disease by segmentation of the hair as well as attributes of an individual (e.g., sex and age). Therefore, an nonenzymatic method of hair digestion and profiling is required. In addition to optimizing and validating a method for measuring the distribution of amino acids in human hair, a robust and comprehensive approach to objectively compare the most effective means of extracting and manipulating chromatographic data to obtain the best limits of detection, linearity, and sensitivity are provided.
Data comparisons were made by operating the mass spectrometer in a mode that rapidly switches between total ion current (TIC) and selected ion monitoring (SIM) modes during each sample injection. In this way, any external confounding factors were negated that may otherwise influence the comparison of the linearity and sensitivity between the two modes of operation. The use of SIM, peak areas, and an internal standard provided significantly better sensitivity and limits of detection than using peak heights, TICs, or no internal standard.
The sample preparation steps included protein acid hydrolysis using hydrochloric acid and trimethylsilyl (TMS) derivatization using N,O-bis(trimethylsilyl)trifluoroacetamide (BSTFA). The optimal derivatization conditions were acetonitrile as reaction solvent, temperature of 100°C, and a reaction time of 30 min.
The method was validated by measuring the amino acid content of myoglobin. This validation was accurate for nine of the fourteen amino acids found in myoglobin and gave detection limits in the range of 0.04–0.1 μmol/L, quantitation limits in the range of 0.1–0.5 μmol/L, recoveries between 80% and 110%, and linear models with coefficients of determination (R2) greater than 0.99 in the tested range from 1 to 300 μmol/L. The remaining five amino acids of myoglobin were deleteriously affected by acid hydrolysis.
Keywords
Amino acid analysis; Protein hydrolysis; Trimethylsilyl (TMS) derivatization; BSTFA; Selected ion monitoring (SIM); GC/MS
Abstract
The distribution of amino acids in hair can divulge information regarding the health (e.g., diabetes) and provide a means for detecting the history of the disease by segmentation of the hair as well as attributes of an individual (e.g., sex and age). Therefore, an nonenzymatic method of hair digestion and profiling is required. In addition to optimizing and validating a method for measuring the distribution of amino acids in human hair, a robust and comprehensive approach to objectively compare the most effective means of extracting and manipulating chromatographic data to obtain the best limits of detection, linearity, and sensitivity are provided.
Data comparisons were made by operating the mass spectrometer in a mode that rapidly switches between total ion current (TIC) and selected ion monitoring (SIM) modes during each sample injection. In this way, any external confounding factors were negated that may otherwise influence the comparison of the linearity and sensitivity between the two modes of operation. The use of SIM, peak areas, and an internal standard provided significantly better sensitivity and limits of detection than using peak heights, TICs, or no internal standard.
The sample preparation steps included protein acid hydrolysis using hydrochloric acid and trimethylsilyl (TMS) derivatization using N,O-bis(trimethylsilyl)trifluoroacetamide (BSTFA). The optimal derivatization conditions were acetonitrile as reaction solvent, temperature of 100°C, and a reaction time of 30 min.
The method was validated by measuring the amino acid content of myoglobin. This validation was accurate for nine of the fourteen amino acids found in myoglobin and gave detection limits in the range of 0.04–0.1 μmol/L, quantitation limits in the range of 0.1–0.5 μmol/L, recoveries between 80% and 110%, and linear models with coefficients of determination (R2) greater than 0.99 in the tested range from 1 to 300 μmol/L. The remaining five amino acids of myoglobin were deleteriously affected by acid hydrolysis.
Introduction
Proteins are an abundant class of large biological molecules common to all life and are the target of a huge number of biological studies. Amino acids are the major constituents of proteins and are also very common targets in biochemical studies. In certain applications, such as in the nutritional content of proteinaceous foodstuffs, it is desirable to understand the relative composition of amino acids for a certain protein or a mixture of proteins. In such applications, it is usually necessary to hydrolyze the proteins using acid hydrolysis until its individual amino acids are released and are available for detection. The most often employed acid for hydrolysis is hydrochloric acid (HCl). The advantage of HCl is that it is relatively non-oxidizing and that it can readily be removed from the hydrolysate. The classical approach of acid hydrolysis is boiling a known amount of proteins in 6 M HCl at 110°C for 18-24 h [1-3]. Following protein hydrolysis, derivatization of the amino acids is usually required for gas chromatographic analysis[4-6].
Most often, amino acid analyzers are used to profile the amino acid distribution of a protein. Commercial analyzers of this kind separate the mixture of amino acids either by ion-exchange chromatography or by high-performance liquid chromatography incorporating post-column derivatization with ninhydrin [4,5,7].but the separation times can be relatively long and detection can be hindered by complex matrix effects, especially from biological fluids like blood, urine, and spinal fluid [8,9]. Analysis using GC typically requires offline pre-column derivatization and is typically more efficient than post-column derivatization [4]. In addition, the proper choice of the derivative can improve volatility, chromatographic performance (e.g., minimizing tailing or adsorption), and improve detector sensitivity [4,10,11]. For GC, many derivatization methods have been reported with success [8,12-15]. For example, free amino acids can be derivatized directly in an aqueous solution using alkyl chloroformates, e.g., methyl, ethyl, or isobutyl. The amino acids react with propyl chloroformate and the derivates can be extracted with an organic solvent for injection directly into the GC/MS [15].
N,O-bis(trimethylsilyl)trifluoroacetamide (BSTFA) is a common silylating reagent that replaces the acidic protons of the amino acids (e.g., SH, OH, NH, and COOH) with nonpolar trimethylsilane (TMS) groups. By replacing the acidic protons with nonpolar TMS groups, the polarity of an amino acid is reduced and its volatility is increased [5,13,14,16-18]. The advantage of TMS derivatization is that it requires a single step whereas other derivatization methods usually have two or more reaction steps [19,20]. The major disadvantages of TMS derivatives are their sensitivity to moisture and the derivatization of Gly and Glu that are affected by the polarity of the solvent [18,20]. For example, both the di-trimethylsilyl derivative and tri-trimethylsilyl derivatives of Gly are obtained after derivatization using either acetonitrile or acetone as solvents [18] and two peaks are observed for Glu when using chloroform as the solvent [20].
The TMS-derivatized amino acids are usually analyzed using gas chromatography/mass spectrometry (GC/MS) and identified according to a combination of their mass spectra and retention times [6,21,22]. GC/MS is a powerful instrument for the separation and identification of components in complex mixtures [23] and plays a central role in amino acid analysis by affording the advantages of improved resolution, sensitivity, and quantification, as well as faster analysis and smaller amounts of sample [1,6,16,24,25].
The previous literature contains inconsistent results about the optimal derivatization conditions of amino acids using BSTFA. These results may be related to many limitations that were observed in previous studies, such as the limited number of replicates for each of the derivatization conditions [26-29], using either peak height or peak area without evaluating the linearity of the calibration, sensitivity, or limit of detection (LOD) [18,19,27], using either TIC or SIM modes without comparing the linearity, sensitivity, or LOD, or using relatively long GC temperature programs (e.g., 54 min [27], 62 min [6], and 90 min [29]).
The use of internal standards is typically required to correct for chemical and analytical losses of amino acids during hydrolysis and derivatization. Kaspar et al. [15] reported that amino acids could be reliably quantified using 19 stable isotope-labeled amino acids as internal standards. The internal isotope standards for each amino acid would be preferable because it typically provides the best reproducibility, best accuracy and most precise result. However, internal isotope standards are very expensive. Acceptable linearity and reproducibility using norvaline (Nor) as internal standard has been reported [11] and Nor is considerably cheaper than deuterated isotope standards for each amino acid. For these reasons, Nor was selected as an internal standard for the present study.
In the present study, the method has been improved, and a comprehensive comparison of the most effective means of manipulating the chromatographic data to optimize the LODs, linearity, and sensitivity is reported. Because the mass spectrometer was operated in “fast automated scan and SIM mode” (FASST) mode, the instrument rapidly switched between TIC and SIM modes during each single injection. This capability removes the effect of any external confounding factors such as instrument drift, carryover or column bleed, which may otherwise influence the comparison of linearity and sensitivity for these two modes of operation. Moreover, this study evaluates the effects of peak area, relative peak area (i.e., referenced to the norvaline internal standard), peak height, and relative peak height (i.e., referenced to the norvaline internal standard) on the calibration figures of merit. The number of trials (9 trials) for each derivatization condition provides confidence in the statistical tests and the time needed to program the GC oven temperature (16 min) is considerably shorter than those reported by others. In addition, the method was reproducible even when data was collected across a span of 60 days.
During the acid hydrolysis step, the majority of the amino acids are unaltered. However, tryptophan (Trp) and cystine (Cyt) are decomposed. Methionine (Met) undergoes oxidation. Glutamine (Gln) and asparagine (Asn) are typically deamidated to aspartic acid (Asp) and glutamic acid (Glu), respectively [1]. In addition, arginine (Arg) decomposes to ornithine during silylation, which can yield inaccurate values in the analysis of free amino acids in extracts of biological fluids as well as cell and tissue extracts [1]. Loosing histidine (His) and Arg amino acids after acid hydrolysis was also reported by Rayner [30]. Therefore, the optimization process evaluated linearity, accuracy, LOD, and limit of quantitation (LOQ) for the 14 remaining amino acids. These amino acids were also selected because they comprise the majority of the amino acids in human hair, which is the intended application for this method. Additionally, these amino acids will be easiest to measure by GC/MS, because they will offer the best signal to noise (S/N) values, and are least affected by co-elution with smaller constituents [29]. Because there is no known hair standard available with which to validate our method, horse heart myoglobin was used to validate the method. Although myoglobin does not contain disulfide bonds, it does contain a high degree of alpha-helical structure, much like the keratins found in hair. The method has been used on dozens of hair samples and has provided adequate within-person variance results when separate aliquots of hair from the same individual are analyzed. Results of the hair analyses are reported in two separate manuscripts, one will submit to the forensic international journal and the second will submit to the diabetes journal.
Materials and Methods
Reagents and Supplies
The 14 amino acids of interest in this study are L-alanine (Ala), L-glycine (Gly), L-Valine (Val), L-leucine (Leu), L-isoleucine (Ile), L-proline (Pro), L-serine (Ser), L-threonine (Thr), L-aspartic acid (Asp), L-glutamic acid (Glu), L-phenylalanine (Phe), L-lysine (Lys), L-tyrosine (Tyr), and L-cystine (Cyt). L-norvaline (Nor) was used as an internal standard. All these amino acid standards were purchased from Sigma-Aldrich (St. Louis, MO, USA). The derivatizing agent was N,O-bis(trimethylsilyl)trifluoroacetamide (BSTFA) (Supelco Analytical, Bellefonte, PA, USA). Hydrochloric acid (HCl) 6 M was used as the hydrolysis agent to liberate free amino acids from horse heart myoglobin. Acetonitrile, methanol, acetone, and chloroform were purchased from GPS Chemicals (Columbus, OH, USA). Horse heart myoglobin was also purchased from Sigma-Aldrich (St. Louis, MO, USA).
This study used 4-mL glass vials with phenolic rubber lined caps (Qorpak, Bridgeville, PA, USA). Solutions were filtered through a 13 mm × 0.45 μm, polyvinylidene difluoride (PVDF) filter (Bonna-Agela Technologies, Wilmington, DE, USA). The nitrogen generator was purchased from Parker Hannifin Corporation (Haverhill, MA, USA).
Standard Stock Solutions
Standard amino acid stock solutions were prepared by dissolving each of the 14 amino acids in 0.1 M HCl to a concentration of 5 μmol/mL. An internal standard stock solution was prepared by dissolving Nor in 0.1 M HCl to a concentration of 10 μmol/mL. The solutions were stored at 4°C until analysis. Calibration standards at 7 different concentrations (0.001-0.3 μmol/mL were prepared using the 14 standard amino acid stock solutions. In each solution, the concentration of internal standard Nor was 0.16 μmol/mL.
Amino Acid Hydrolysis
Determining the amino acid content of myoglobin involved the acid hydrolysis in 6 M HCl for 24 h until the protein-bound amino acids were released and were available for detection. The myoglobin sample was separated into three subsamples and precisely weighed to approximately 3.0 mg and transferred into a 4-mL glass vial with a phenolic rubber lined cap. Thereafter, 1.0 mg of Nor was added. Each subsample was hydrolyzed by adding 0.3 mL of 6M HCl to the vial and was tightly capped. To heat the samples, an aluminum block with holes to accommodate the glass vials was mounted on a hot-plate for 24 h at 110°C. After cooling the solution to room temperature, the solutions were filtered and dried under a constant 4 L/min nitrogen stream.
Amino Acid Derivatization
4.4.1) Effect of Solvent: A 100 μL aliquot of the 0.33 μmol/mL standard amino acid solution was pipetted into a 4-mL glass vial and dried under a nitrogen stream. The existence of moisture can result in poor reaction yield and instability of the derivatized products. The dried amino acid residues were dissolved in a 100-μL aliquot of acetonitrile and derivatized with the addition of a 100-μL aliquot of BSTFA. The glass vial was tightly capped, ultrasonicated for 1 min, and mounted on a hot-plate for 30 min at 100°C. For the solvent study, the only change made was to substitute the acetonitrile with chloroform and acetone. Triplicate samples for each solvent were prepared and replicated three times to yield 9 measurements for each solvent.
Effect of Time and Temperature Reactions
Standard solutions of 0.33 μmol/mL were prepared and stored at 4 °C. A 100-μL aliquot of this solution was transferred into a 4-mL glass vial and dried under constant nitrogen stream. The dried amino acid residues were dissolved in a 100-μL aliquot of acetonitrile and derivatized with the addition of a 100-μL aliquot of BSTFA. The glass vial was tightly capped, ultrasonicated for 1 min and mounted on a hot-plate at 50, 100, or 150°C with reaction times of 15, 30, 45, or 60 min, separately. Precision was measured by using Triplicate samples for each time at each temperature were prepared and replicated three times to yield 9 measurements for each time at each temperature.
GC/MS Analysis
Amino acid derivatives were analyzed with a Shimadzu GC/MS instrument (QP-2010SE, Scientific Instrument, Inc. Columbia, MA, USA). Amino acids were separated on 5% diphenyl-dimethylpolysiloxane capillary column (SHRX1-5MS, 30 m X 0.25 mm X 0.25 μm). Ultra-high purity helium was used as a carrier gas at a constant flow rate of 21.5 mL/min. The transfer column flow was 1.0 mL/min.
The temperature of the column was programmed to rise from 70°C to 170°C at a rate of 10°C/min, and then was ramped to 280°C at a rate of 30°C/min, at which it was held for 3 min. The total run time was 16.6 min. Sample injection was performed in split injection mode (1:20 ratio) at 280°C using an injection volume of 1 μL. Triplicates injections for each sample were run in a random block design (i.e., complete set of samples analyzed in randomized order, before the next set of sample replicates are analyzed in a rerandomized order) using acetonitrile as a solvent blank before each injection. Three solvent vials of acetonitrile, methanol, and acetone were used sequentially as cleaning solvents for the autosampler injection syringe.
The mass spectrometer was operated in fast automated scan and SIM type (FASST) mode, which switches between full scan mode and selected ion monitoring during a single analysis. The mass spectrometer was operated in full scan mode (TIC) from m/z of 50 to m/z of 500 with a scan time of 0.3 s, and selected ion monitoring mode (SIM) with a scan time of 0.2 s. For SIM, the appropriate ion set of one target ion and two reference ions as characteristics mass fragments of the derivatized amino acid was used. Selection of the characteristic ions for the quantification ion is very important. The target ions are the fragment ions of the highest intensity (base peak), while the reference ions are two other fragment ions of the next greatest intensities [27]. Table 1 presents the target and reference ions that were quantify as a set of each amino acid.
| Amino acid | Target ion (m/z) | Reference ions (m/z) |
|
Ala |
116 |
73, 147 |
|
Gly of di-trimethylsilyl derivative |
102 |
73, 147 |
|
Val |
144 |
73, 218 |
|
Nor |
144 |
73, 218 |
|
Leu |
158 |
73, 102 |
|
Ile |
158 |
73, 218 |
|
Pro |
142 |
73, 216 |
|
Gly of tri-trimethylsilyl derivative |
174 |
73, 248 |
|
Ser |
304 |
218, 73 |
|
Thr |
73 |
218, 291 |
|
Asp |
232 |
156, 73 |
|
Glu |
246 |
73, 128 |
|
Phe |
218 |
192, 73 |
|
Lys |
174 |
156, 73 |
|
Tyr |
218
|
179, 280
|
|
Cyt |
218 |
147, 73 |
Table 1. Target and reference ions for SIM mode as characteristics mass fragments of the 14 amino acids.
Acid Hydrolysis Effects on Free Amino Acids
Triplicate standard amino acid solutions were prepared by dissolving approximately 0.1 mg of each amino acid and Nor in 1 mL of 0.1 M HCl. The aqueous mixture was split into two glass vials and dried under a constant nitrogen stream. One half was derivatized and analyzed by GC/MS, whereas the other half was hydrolyzed using 1 mL of 6 M HCl, derivatized, and analyzed.
Calibration Curves and Reproducibility
To evaluate the linearity and sensitivity of the signal with respect to concentration, eight linear calibrations were generated for each amino acid. The first set of calibrations modeled the absolute peak areas of the corresponding amino acids with respect to their amino acid concentrations (μmol/mL). The second set of calibrations modeled the relative peak areas referenced to the Nor internal standard. The third set of calibrations modeled the absolute peak heights with respect to the amino acid concentrations. The fourth set of calibrations modeled the relative peak heights referenced to the Nor internal standard. These four calibrations were obtained from the total ion chromatograms and the extracted ion chromatograms (using the target/quant ions reported in the section above) to yield a total of eight calibrations.
Data Analysis
Statistical analyses were performed using MATLAB R2012b or R2014a (MathWorks, Natick, MA). Excel 2010 (Microsoft, Redmond, WA) was also used to calculate the concentrations and calibrations. All statistical tests were conducted at the 95% confidence level.
Results and Discussion
Method validation included the evaluation of the independent procedures of acid hydrolysis, amino acid derivatization, and GC/MS analysis. To fully assess the performance of these procedures, a standard amino acid mixture enabled the verification of the GC/MS method, including derivatization and acid hydrolysis effects on amino acids, while the reference protein sample (i.e., horse heart myoglobin) helped assess the effectiveness of acid hydrolysis. In addition, any variation in conditions from one run to the next was controlled by referencing all data to the Nor internal standard peak [5,11,15,31].
Effect of Solvent on Derivatization
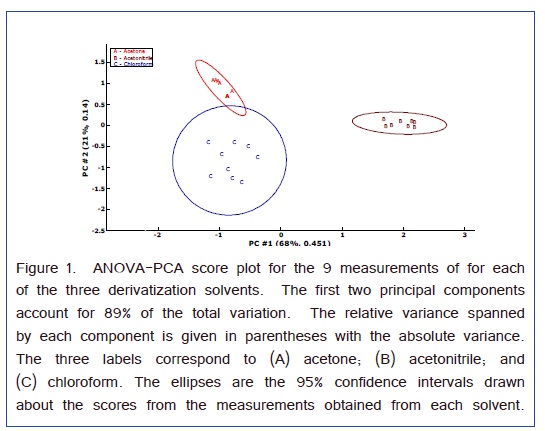
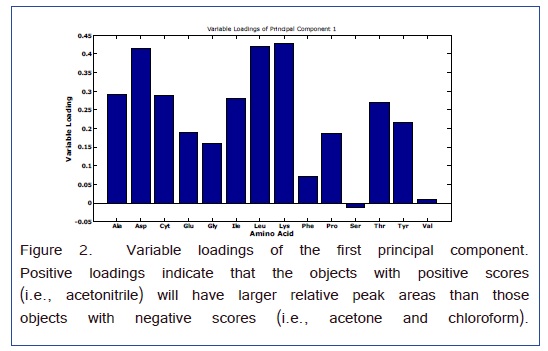
Analysis of variance-principal component analysis (ANOVA-PCA) evaluated the sources of variation arising from the differences among the relative peak areas (i.e., referenced to the Nor internal standard) of the amino acids and the three different solvents that were used for derivatization [32]. As presented in Figure 1, the 3 solvents formed well resolved clusters of principal component scores. The ellipses around the scores are 95% CI and the effects of all three solvents were significant. The first two principal components account for 89% of the total variance. The first principal component separated the acetonitrile from the other two solvents. Hence, the variable loadings of this principal component will indicate the overall effect of the solvent. Figure 2 are the variable loadings of the first component; positive loading correspond to amino acid peaks that are larger for the acetonitrile solvent and negative loadings correspond to amino acid peaks that are larger for one of the other two solvents.
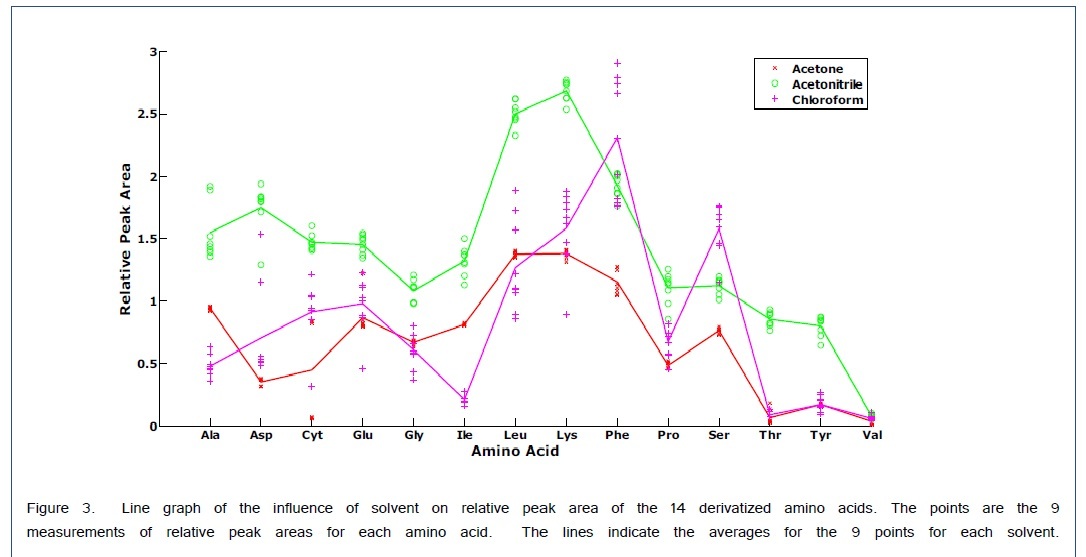
A scatter plot with the superimposed averages gives the relative peak areas for each amino acid with respect to the solvent in Figure 3. This graph is consistent with the variable loadings plot in that chloroform gives a larger Ser peak area than that obtained from acetonitrile. For the Phe peak, although the average is larger for chloroform the relative peak areas are much less precise which gave a lower weight for the variable loading in Figure 2. Therefore, the Phe variable loading on the first PC is positive indicating a favorable result for acetonitrile as a solvent. For all the other amino acids, acetonitrile appears to work the best.
The derivatization of Gly and Glu were affected by the polarity of the solvent. Sharp single peaks were obtained for all amino acids except Gly, in which case both the di-trimethylsilyl derivative and tri-trimethylsilyl derivative were obtained after derivatization using acetonitrile or acetone as solvents.In contrast to these solvents, two peaks were observed for Glu when using chloroform as the solvent. Ala, Val, Nor, Leu, Ile, Pro, and Phe produced di-trimethylsilyl derivatives in all solvents and Ser, Thr, Asp, Glu, Lys, and Tyr yielded tri-trimethylsilyl derivatives in all solvents. Cyt formed a tetra-trimethylsilyl derivative. These results are similar to those obtained by Cehrke et al. [20] who also found that the derivatization of Gly and Glu were affected by the polarity of the solvent. For less polar solvents such as chloroform, only one peak was obtained for glycine and two peaks were obtained for Glu. Conversely with more polar solvents such as acetonitrile and acetone, one observes two peaks for Gly and only one peak for Glu. The observation of two products for certain amino acids, although not ideal, does not necessarily prevent quantitation. If both products are chromatographically resolved, the sum of the two peak heights or peak areas can be used for quantitation.
Effect of Temperature and Time on Derivatization
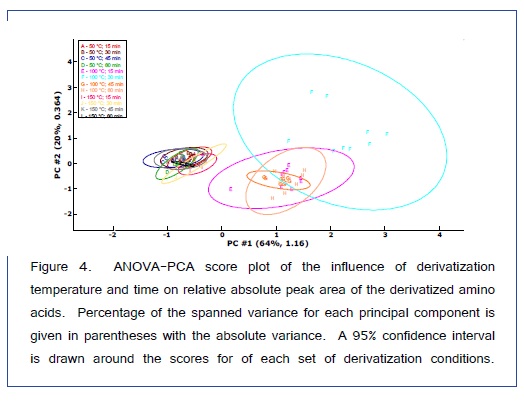
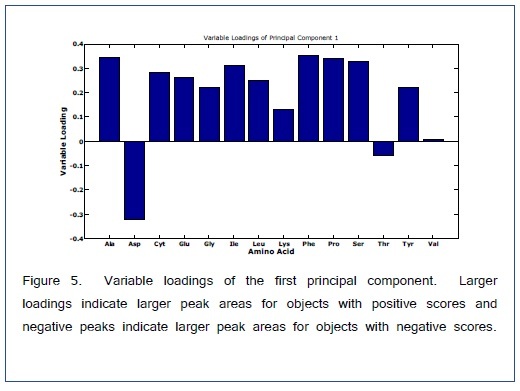
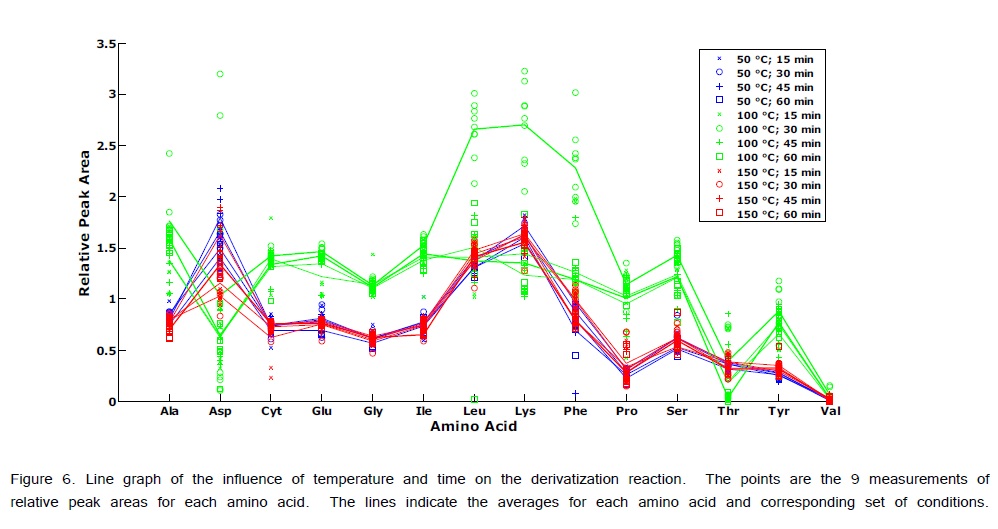
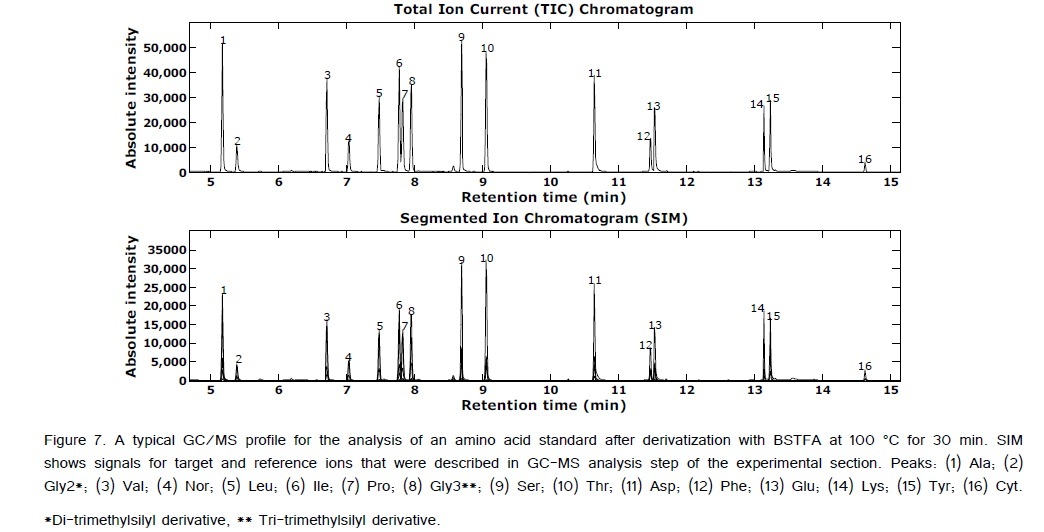
ANOVA-PCA explored the effects of different temperatures (50, 100, and 150°C) and different times (15, 30, 45, and 60 min) on the relative peak areas for the 14 amino acid derivatives. Figure 4 is the PCA score plot, all scores for the reaction temperature of 100°C are positive on the first principal component. The most extreme cluster of scores is for 100°C and 30 min. The first two principal components account for 84% of the total variance. The variable loadings of the first component (Figure 5) and the scatter plot in Figure 6, the trend of all amino acids are positively enhanced using the reaction temperature of 100°C for 30 min, except for Asp and Thr amino acids Consequently, the reaction temperature of 100 °C for 30 min has the largest product yield. This result is in agreement with Shen et al. [24] and Deng et al. [33] who concluded that the optimal derivatization conditions were acetonitrile as a solvent, a temperature of 100 °C, and a reaction time of 30 min. Figure 7 is a typical GC/MS profile for the standard amino acid mixture at a concentration of 0.3 μmol/mL that was obtained under these optimal conditions.
Total ion Current Mode (TIC) and Selected Ion Monitoring Mode (SIM)
Paired sample t-tests were conducted to compare SIM and TIC modes using different parameters: the coefficient of determination (R2) and the limit of detection (LOD) of the calibration line. The concentration LOD is the lowest concentration of an analyte in a sample that can be detected above the backgrounds, but not necessarily quantified [34]. The LOD was calculated by dividing three times the standard error of the calibration line at the intercept by the slope of the calibration line [34]. First, the absolute peak areas and the relative peak areas (i.e., referenced to the Nor internal standard) for each mode (SIM or TIC) were compared separately. The R2 and LOD values were significantly different between the absolute and relative peak area calibrations for both SIM and TIC acquisition modes. The absolute and relative peak heights (i.e., referenced to the Nor internal standard) for each analysis mode were also compared.
The R2 and LOD values were significantly different between the absolute and relative peak height calibrations. Based on these results, the relative peak area and the relative peak heights are preferred to the absolute values for both SIM and TIC modes of data acquisition, as presented in Table 2. R2 and LOD are the average of R2 and LODs of the 14 amino acids. Moreover, the minimum and maximum values of LOD indicate that the relative values are always more sensitive than the absolute values. Second, the relative peak area (i.e., referenced to the Nor internal standard) and the relative peak heights (i.e., referenced to the Nor internal standard) were compared between the two acquisition modes (Table 3). The significant difference noted in relative peak area. The R2 and LOD in SIM mode of relative peak area is better than TIC mode, as well as the lower minimum and maximum values.
| Calibration parameters | SIM (n=14) | TIC (n=14) | ||||||||||
| Absolute Area | Relative Area | P-value | Absolute Height | Relative Height | P-value | Absolute Area | Relative Area | P-value | Absolute Height | Relative Height | P-value | |
| Mean ± 95%CI | Mean ± 95%CI | Mean ± 95%CI | Mean ± 95%CI | Mean ± 95%CI | Mean ± 95%CI | Mean ± 95%CI | Mean ± 95%CI | |||||
|
R2 |
0.95 |
0.993 |
0.001 |
0.94 |
0.97 |
0.002 |
0.95 |
0.97 |
0.01 |
0.95 |
0.97 |
0.02 |
|
± 0.02 |
± 0.003 |
± 0.03 |
± 0.01 |
± 0.03 |
± 0.02 |
± 0.01 |
± 0.02 |
|||||
|
LOD (µmol/L) |
0.09 ± 0.04 |
0.06 ± 0.02 |
0.04 |
0.13 ± 0.09 |
0.07 ± 0.01 |
0.03 |
0.14 ± 0.08 |
0.07 ± 0.01 |
0.01 |
0.09 ± 0.04 |
0.07 ± 0.02 |
0.04 |
|
Min LOD |
0.05 |
0.04 |
0.05 |
0.05 |
0.06 |
0.07 |
0.08 |
0.07 |
||||
|
Max LOD |
0.3 |
0.1 |
0.9 |
0.1 |
0.9 |
0.1 |
0.9 |
0.1 |
||||
Table 2. The comparison of absolute peak area, relative peak area, absolute peak height, and relative peak height in SIM and TIC modes, the mean is the average of the 14 amino acids. A matched-sample t-test was used to determine the p-values.
| Calibration parameters | SIM | TIC | p-values |
| Mean ± 95% CI (n=14) | Mean ± 95% CI (n=14) | ||
|
R² of relative peak area |
0.993 ± 0.003 |
0.97 ± 0.02 |
0.04 |
|
LOD of relative peak area |
0.06 ± 0.02 |
0.07 ± 0.01 |
0.05 |
|
Min LOD |
0.04 |
0.07 |
|
|
Max LOD |
0.1 |
0.1 |
|
|
R² of relative peak height |
0.97 ± 0.01 |
0.97 ± 0.02 |
0.93 |
|
LOD of relative peak height |
0.07 ± 0.03 |
0.07 ± 0.01 |
0.4 |
|
Min |
0.05 |
0.07 |
|
|
Max |
0.1 |
0.1 |
Table 3. Comparison of average R2 values and LOD values of relative peak area and relative peak height of the 14 amino acids in SIM and TIC modes. A matched sample t-test was used to determine the p-values.
Third, paired sample t-tests were also conducted to compare R2 of relative peak areas and relative peak heights within the SIM mode. The R2 was significantly larger for relative peak area than for relative peak height (p= 0.02). According to these results, the coefficient of determination of the relative peak area is better than that of the relative peak height.
To summarize, the internal standard (Nor) improved the coefficient of determination for both MS modes, and peak area provided better performance than peak height for the TIC and SIM modes. The SIM mode had better sensitivity and linearity than the TIC mode. The best method was therefore SIM mode, with the peak areas normalized to the internal standard, Nor. The calibration curve results for the relative peak areas for the 14 amino acids studied are reported in Table 4.
| Retention time (min) | Regression line (n=3) | Limit of detection (µmol/L) | Limit of | |||
| Amino acid | quantitation | |||||
| R2 ± 95% CI | Slope ± 95% CI | Intercept ± 95% CI | (µmol/L) | |||
|
Ala |
5.19 |
0.991 ± 0.003 |
5.9 ± 0.2 |
0.042 ± 0.007 |
0.07 |
0.3 |
|
Gly |
5.36 |
0.99 ± 0.01 |
8.7 ± 0.7 |
0.04 ± 0.04 |
0.04 |
0.2 |
|
Val |
6.73 |
0.991 ± 0.003 |
6.9 ± 0.2 |
0.031 ± 0.005 |
0.04 |
0.2 |
|
Leu |
7.85 |
0.994 ± 0.003 |
5.3 ± 0.1 |
0.03 ± 0.02 |
0.07 |
0.2 |
|
Ile |
7.79 |
0.993 ± 0.002 |
6.5 ± 0.2 |
0.031 ± 0.005 |
0.04 |
0.1 |
|
Pro |
7.84 |
0.995 ± 0.002 |
8.5 ± 0.1 |
0.021 ± 0.009 |
0.04 |
0.1 |
|
Ser |
8.71 |
0.996 ± 0.009 |
9.8 ± 0.2 |
0.008 ± 0.007 |
0.04 |
0.1 |
|
Thr |
9.07 |
0.993 ± 0.006 |
12.7 ± 0.7 |
0.04 ± 0.09 |
0.08 |
0.2 |
|
Asp |
10.66 |
0.997 ± 0.002 |
15.9 ± 0.9 |
-0.09 ± 0.07 |
0.05 |
0.1 |
|
Glu |
11.48 |
0.99 ± 0.01 |
5.03 ± 1.27 |
-0.176 ± 0.007 |
0.04 |
0.3 |
|
Phe |
11.54 |
0.996 ± 0.009 |
9.6 ± 0.7 |
-0.07 ± 0.06 |
0.07 |
0.2 |
|
Lys |
13.15 |
0.99 ± 0.02 |
4.5 ± 0.3 |
-0.11± 0.01 |
0.08 |
0.2 |
|
Tyr |
13.24 |
0.995 ± 0.003 |
9.5 ± 0.5 |
-0.24 ± 0.09 |
0.08 |
0.2 |
|
Cyt |
14.64 |
0.97 ± 0.01 |
1.3 ± 0.4 |
-0.2 ± 0.1 |
0.1 |
0.5 |
Table 4. Retention time, R2 values, calibration equation, limit of detection (LOD), and limit of quantitation (LOQ) of the derivatized amino acids using SIM mode after normalization to the internal standard, Nor.
Limits of Detection and Quantitation
The limit of quantitation (LOQ) is defined as the concentration that gives a signal to noise (S/N) ratio of 10 [11,15] whereas the LOD is usually defined as the concentration at which the signal to noise ratio is 3 [11,15,24]. In Table 4, the LODs were calculated by dividing three times the standard error of the calibration line at the intercept by the slope of the calibration line and the LOQs were calculated by dividing 10 times the standard error of the calibration line at the intercept by the slope of the calibration line. As tabulated in Table 4, for liquid standard injections, the LODs and the LOQs were in the ranges of 0.04-0.1 μmol/L and 0.1-0.5 μmol/L, respectively, depending on the amino acid under consideration.
Bartolomeo et al. [11] reported less sensitive LOQs than those reported in current method (3-16 μmol/L) for BSTFA-derivatized amino acids. Kasper et al. [15] also reported less sensitive LOQs of 0.3-30 μmol/L and LODs (0.03-0.12 μmol/L) for propyl chloroformate-derivatized free amino acids in biological fluids. Whereas, Shen et al.[24] reported similar LODs of 0.08-0.7 μmol/L for BSTFA-derivatized amino acids.
Acid Hydrolysis Effects on Free Amino Acids
Acid hydrolysis is a crucial step that influences amino-acid recovery. During acid hydrolysis, most (nine) of the amino acids were unaffected. However, Gln and Asn are typically deamidated to Asp and Glu, respectively. Arg decomposes to ornithine during silylation, which can yield inaccurate values in the analysis of free amino acids in extracts of biological fluids as well as cell and tissue extracts. Cyt totally decomposed. Ser, Tyr, and Thr were also partially lost due to their lability during acid hydrolysis [2,3,25].
The calculated recoveries were based on the measured weight of the amino acids in the starting solutions and the calculated mass based from the relative peak areas. To calculate the original amino acid mass from the peak area, external calibrations were used liquid standards, the known GC split, the known injection volume, and solvent dilution factors. The liquid calibration modeled the relative peak area of each amino acid with respect to the corresponding prepared concentrations. Amino acid standard in Table 5 reports the amino acids that have been recovered after acid hydrolysis.
| Amino acid | Actual weight (mg) | Calculated weight via deriv. GC/MS (mg) | Recovery (%) | Calculated weight after acid hydrolysis and deriv. GC/MS (mg) | Recovery (%) |
| Mean ± 95% CI | Mean ± 95% CI | Mean ± 95% CI | Mean ± 95% CI | Mean ± 95% CI | |
| (n=3) | (n=9) | (n=9) | (n=9) | (n=9) | |
|
Ala* |
0.12 ± 0.01 |
0.14 ± 0.02 |
118 ± 2 |
0.13± 0.03 |
105* ± 2 |
|
Gly |
0.13 ± 0.02 |
0.12 ± 0.02 |
97 ± 2 |
0.07 ± 0.01 |
72 ± 1 |
|
Val* |
0.078 ±0.004 |
0.08 ± 0.01 |
110 ± 1 |
0.08 ± 0.01 |
109* ± 7 |
|
Leu* |
0.147 ±0.002 |
0.14 ± 0.01 |
93 ± 6 |
0.16 ± 0.02 |
105* ± 3 |
|
Ile* |
0.19 ± 0.02 |
0.17 ± 0.03 |
89 ± 3 |
0.18 ± 0.04 |
88* ± 3 |
|
Pro* |
0.16 ± 0.02 |
0.14 ± 0.02 |
99 ± 4 |
0.15 ± 0.02 |
108* ± 1 |
|
Ser |
0.16 ± 0.03 |
0.18 ± 0.04 |
103 ± 4 |
0.12 ± 0.04 |
70 ± 4 |
|
Thr |
0.21 ± 0.04 |
0.21 ± 0.03 |
98 ± 2 |
0.15 ± 0.03 |
75 ± 2 |
|
Asp* |
0.16 ± 0.03 |
0.16 ± 0.04 |
105 ± 3 |
0.19 ± 0.02 |
103* ± 1 |
|
Glu* |
0. 18 ± 0.02 |
0.21± 0.04 |
106 ± 2 |
0.16 ± 0.03 |
92* ± 1 |
|
Phe* |
0.22 ± 0.01 |
0.22 ± 0.03 |
103 ± 1 |
0.19 ± 0.04 |
91* ± 2 |
|
Lys* |
0.18 ± 0.02 |
0.21 ± 0.02 |
111 ± 1 |
0.13 ± 0.01 |
93* ± 5 |
|
Tyr |
0.21 ± 0.02 |
0.23 ±0.02 |
110 ± 1 |
0.14 ± 0.1 |
73 ± 4 |
|
Cyt |
0.29 ± 0.03 |
0.3 ± 0.1 |
102 ± 3 |
||
|
Average recovery (%) |
103 |
99* |
* Average of the best-recovered amino acids: Ala, Leu, Phe, Asp, Glu, Val, Ile, Pro, and Lys.
Table 5. Amino acid recovery (%) calculated from single amino acids.
Recoveries after the acid hydrolysis step were 70, 74, and 75% for Ser, Tyr, and Thr, respectively. As discussed above, Gly is known to give two derivatives after the BSTFA reaction in polar solvents, so was not included in the acid-hydrolysis validation study. Therefore, the validation parameters were estimated using the nine best-recovered amino acids: Ala, Leu, Phe, Asp, Glu, Val, Ile, Pro, and Lys. These 9 amino acids all were recovered within the accepted range 80-110% [35]. Recoveries of the best amino acids after acid hydrolysis ranged from 88.9% for Ile up to 109.8% for Val, while the average recovery was 99.7%, which is within 80-110% that is widely considered acceptable [35].
Accuracy
A myoglobin standard with a defined distribution of amino acids was hydrolyzed and subjected to the analysis. This evaluation was important because amino acids differ in the ease with which they are liberated from peptide linkages during acid hydrolysis. Amino acids such as Val and Ile are linked by peptides bonds that are not easily broken [2,3,25,36]. Val and Ile values were lower than expected because of incomplete hydrolysis; peptides with an Ile-Val pair in their sequence are known to be resistant to acid hydrolysis because of steric effects [2].
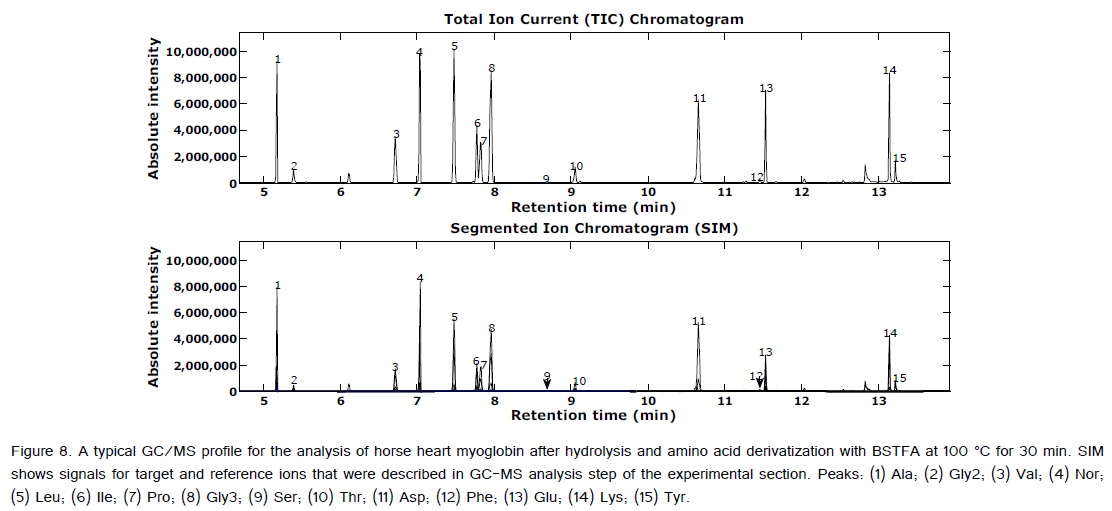
Table 6 reports the results of the myoglobin experiment. As expected, Cyt appeared to be completely hydrolyzed following acid hydrolysis [2,3,11,25]. Ser, Tyr, Pro, and Thr are completely liberated after hydrolysis for 20 h, but they progressively decompose under acid hydrolysis [2,3,11,25,31]. Moreover, because Gln and Asn were converted to Glu and Asp [31], Table 6 reports the actual and measured Asp mole fractions. The measured mole fractions for myoglobin Asp was obtained from sum of Asp and Asn. The sum of the mole fractions for Glu and Gln are also reported. Additionally, based on the acid hydrolysis effects on free amino acids presented in Table 5, the validation parameters were estimated using the best-recovered amino acids: Ala, Val, Leu, Ile, Phe, Asp, Glu, Pro, and Lys. The TIC and SIM Chromatograms of the amino acids obtained from myoglobin are given in Figure 8.
Based on these results, the relative peak areas of the SIM mode gave the better performance. The measured mole fractions of myoglobin amino acids were obtained by: (1) the concentration (μmol/mL) for each amino acid was calculated using external calibration curves of liquid standards, (2) the amino acid concentrations from the calibration curves were used to calculate the moles for each amino acid, (3) the total moles of analyzed amino acids were calculated, (4) the molar ratios were calculated by dividing the measured moles for each amino acid by the total experimental moles, and (5) multiplying fraction of the amino acid by 100 expresses the mole fraction as a percentage.
| Amino acid | True mole percent obtained from sequence | Experimently obtained mole percent | Absolute error | True mole percent of the nine best recovered amino acids obtained from sequence | Experimentaly obtained | Absolute error (mole percent) |
| Mean ± 95% CI | mole percent of the nine best recovered amino acids# | |||||
| n=9 | Mean ± 95% CI | |||||
| n=9 | ||||||
|
Ala |
11.2 |
13 ± 1 |
2.2 |
14.2 |
15 ± 1 |
0.8 |
|
Gly |
11.2 |
11 ± 1 |
-0.2 |
|||
|
Val |
5.1 |
3.4 ± 0.3 |
-1.7 |
6.5 |
5.1 ± 0.3 |
-1.4 |
|
Leu |
12.5 |
15 ± 1 |
2.5 |
15.8 |
18 ± 1 |
2.2 |
|
Ile |
6.6 |
6 ± 1 |
-0.6 |
8.4 |
7 ± 1 |
-1.4 |
|
Pro |
2.9 |
4 ± 1 |
1.1 |
3.7 |
4.3 ± 0.2 |
0.6 |
|
Ser |
3.7 |
1.1 ± 0.1 |
-2.6 |
|||
|
Thr |
5.1 |
2.4 ± 0.1 |
-2.7 |
|||
|
Asp* |
7.4 |
9 ± 1 |
1.6 |
9.3 |
11 ± 1 |
1.7 |
|
Glu** |
13.9 |
15 ± 1 |
1.1 |
17.8 |
18 ± 1 |
0.2 |
|
Phe |
5.1 |
5.4 ± 0.4 |
0.3 |
6.5 |
6.4 ± 0.4 |
-0.1 |
|
Lys |
13.9 |
13.1 ± 0.4 |
-0.8 |
17.8 |
18 ± 1 |
0.2 |
|
Tyr |
1.4 |
0.4 ± 0.2 |
-1 |
|||
|
Total |
100 |
100 |
-0.8 |
100 |
100 |
2.8 |
* Asp is the sum of Asn and Asp.
** Glu is the sum of Gln and Glu.
# The best-recovered amino acids: Ala, Leu, Phe, Asp, Glu, Val, Ile, Pro, and Lys.
Table 6. Measured mole percentage of amino acid composition for myoglobin protein and true mole percentage of amino acids obtained from the sequence.
Conclusion
The presented method was successfully developed for the determination of amino acids in protein. BSTFA derivatization with GC/MS analysis offered a balanced compromise of features over other approaches of amino acid analysis. The derivatization is a single step procedure, the total GC/MS analysis time is 16.6 min, and the MS detection software enabled FASST mode switching between full scan mode and selected ion monitoring mode during a single analysis. Real-time switched-mode data acquisition enabled a direct comparison between the different modes of operation (SIM versus TIC). Peak areas provided lower LODs than peak heights for TIC and SIM modes. The SIM mode had better LODs than the TIC mode, as expected. The best method was therefore SIM mode with the peak area normalized to the norvaline internal standard. The analytical procedures demonstrated in this paper are expected to be applicable to other proteins, and have been reproducible with hair analysis [29].
In addition, ANOVA-PCA was applied for the first time for optimizing the derivatization of a complex set of samples. This method can simplify the optimization when there are many response variables.
Acknowledgements
Dr. Zhengfang Wang and Dr. An Yan are acknowledged for their help.
References
1. Blackburn S (1968) Amino Acid Determination Methods and Techniques, Marcel Dekker Inc., New York.
4. Knapp DR (1979) Handbook of Analytical Derivatization Reaction, Wiley & Sons, New York, NY.
8. Campbell MK, Farrell SO (2006) Biochemistry, Thomson Brooks/Cole, Belmont, CA.
10. Anders JC (2002) Advances in Amino Acid Analysis. BioPharm Int 4:32-39.
16. Gross J (2004) Mass Spectrometry, Springer, New York, NY.
17. Miller J (2005) Chromatography: Concepts and Contrasts, John Wiley & Sons Inc., Hoboken, NJ.
22. Berezin VG (1996) Capillary Gas-Solid Chromatography. Usp Khim 65: 991-1012.
36. Hunt S (1985) Chemistry and Biochemistry of the Amino Acids, Chapman and Hall, London.